|
|
Université catholique
de Louvain (UCL-Bruxelles)
Louvain Drug Research
Institute > Cellular and Molecular Pharmacology |
|
|
Novel
antibiotic targets and drug design |
|
Quick
links
|
World-wide increase
of bacterial resistance to antibiotics makes essential the discovery of
new agents directed against unexploited bacterial targets.
In this context,
we analyse natural mutants with impaired growing capabilities to detect
genes products having a character of essentiality. We then undertake
the design or proactive selection of inhibitors of the function of these
proteins.
These inhibitors
could represent new classes of antibiotics.
These
research programs are also linked to those exploring drug-membrane
interactions and the chemotherapy
of intracellular infection.
|
|
|
|
|
|
|
|
|
| |
Team
- Senior investigators:
F. Van Bambeke,
P.M. Tulkens, J. Poupaert
and R. Frederick*
- PhD student:
A. Ameryckx
- Former investigators:
M. Renard* (2000-2002),
P. Jacquemin* (2002), D. Van
Ackeren (2002-2004), E. Colacino* (2003-2004), C.
Dax* (2003-2004),
I. Tytgat (2004-2008)
* joint programme with the group of Pharmaceutical
Chemistry
Collaborations
- M. Prevost and M. Rooman (Structure
et Fonction des Membranes biologiques, Université libre de Bruxelles,
Brussels, Belgium)
- C. Duez
and B. Joris (Centre d'ingénierie
des protéines, Université de Liège, Liège)
- P.
Courvalin (Unité
des Agents antibactériens, Institut Pasteur, Paris, France)
- C. Franchini (Department
of Pharmacochemistry, University of Bari, Italy)
- P.
Taylor (School of Pharmacy,
University of London, London, UK)
Main research programs
Inhibitors
of D-Ala-D-Ala ligases
The D-alanine:D-alanine
(D-Ala:D-Ala) ligase, which catalyses dimerization of D-alanine before its
incorporation in late peptidoglycan precursors, is essential for bacteria
since natural mutants having lost activity are incapable of growing unless
rescued by the addition of D-Ala:D-Ala in their culture medium. The
reaction catalysed by the D-Ala:D-Ala ligase involves the formation of D-alanylphosphate
as a tetrahedral intermediate.
The enzyme is strongly inhibited by
D-cycloserine and methylphosphinophosphate but none of these inhibitors fulfil
the requirements for becoming useful drugs.
In collaboration with the 'Université
libre de Bruxelles', we have characterized the active site of the enzyme
by 3-D computer-aided modelling (Figure 1). The model successfully explained
the inactivity of four spontaneous mutants, including those which could not
be explained by inspection of the published X-ray structures of this enzyme.
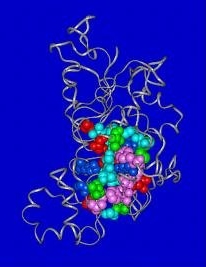 |
Figure 1:
3D-model ( tube diagram)
of the D-Ala:D-Ala ligase of E. faecalis using the X-ray
structure of the Escherichia coli enzyme complexed with ADP
and the methylphosphinophosphate inhibitor as a template.
The image highlights the residues (depicted as balls) found in contact
with ADP and phosphinophosphate.
Color code of residues:
- green, hydrophobic;
- pink, aromatic;
- dark blue, basic;
- light blue, polar;
- red, acidic.
From Prevost et al.,
2000 |
|
Inhibitors with drugable properties are now looked for using (i) rational,
target-based approaches, (ii) empirical pharmacochemical, structure-based
approaches starting from known inhibitors, (iii) screening of natural sources.
These novel compounds are tested for
their capacity to inhibit the enzyme (using a purified protein) as well as
the bacterial growth (using clinical isolates of important human pathogens).
A first successful example
is represented by benzoxazoles, which were designed de novo starting
from computational modeling (Figure 2).
|
|
|
Figure 2:
Left:
Docked pose of the most active benzoxazole (IT16) in the E. coli
D-Ala-D-Ala ligase enzymatic pocket. IT16 is depicted as ball-and-sticks
and interacting residues and ATP are represented as thick lines.
The two magnesium ions are depicted as pink spheres. Oxygens, nitrogens,
carbons, phosphorus and hydrogens are colored in red, blue, green,
magenta and white respectively
Right: chemical structure, inhibitory activity towards
E. faecalis D-Ala-D-Ala ligase, scoring in computational analysis,
and antimicrobial activity of benzoxazole compounds.
From Tytgat
et al., 2008
|
|
Selected References on
D-Ala-D-Ala ligases and inhibitors thereof (by
reverse chronological order; for a full reference list, see our publication
list)
- Tytgat I, Colacino E,
Tulkens PM, Poupaert JH, Prévost M, Van Bambeke F. 2009. DD-Ligases
as a Potential Target for Antibiotics: Past, Present and Future. Current Medicinal
Chemistry 16: 2566-2580. Review (PDF)
- Tytgat
I, Vandevuer S, Ortmans I, Sirockin F, Colacino E, Van Bambeke F, Duez C,
Poupaert JH, Tulkens PM, Dejaegere A, Prévost M. 2009. Structure-based
design of benzoxazoles as new inhibitors for D-alanyl - D-alanine ligase.
QSAR and Combinatorial Sciences 28: 1394-1404. (PDF)
- Poupaert J, Prevost M.,
Vandevuer S, Van Bambeke F, Colacino E, Tytgat I, Tulkens PM. Inhibitors of
D-Ala-D-Ala-ligase as antibacterial agents. Patent number: WO2009080788 (A2);
classification: international: C07D263/56; C07D263/00; european: C07D263/56.
Application number: WO2008EP68100 20081219; Priority number(s): EP20070150355
20071221 (PDF)
- Gholizadeh Y, Prévost
M, Van Bambeke F, Casadewall B, Tulkens PM, Courvalin P. 2001. Sequencing
of the ddl Gene and Modeling of the mutated D-alanine:D-alanine Ligase in
Glycopeptide-Dependent Enterococcus faecium. Prot. Sci. (2001) 10: 836-844.
(PDF)
- Prévost
M, Van Belle D, Tulkens PM, Courvalin P, Van Bambeke F. 2000. Modeling of
Enterococcus faecalis D-alanine:D-alanine ligase: structure based study of
the active site in the wild-type enzyme and in glycopeptide-dependent mutant.
J. Mol. Microbiol. Biotech. 2:321-330 (PDF)
Other
targets
Other targets are being
actively explored. Three orientations are being followed, namely
- the evaluation of antibiotics
acting on the bacterial envelope, for which we explore their ability to
specifically destabilize the phospholipidic membrane (this work is described
in more details in the section drug-membrane
interactions) in relation with their enhanced bactericidal efficacy
(Figure 3A);
- novel benzothiazole
derivatives for which the mode of action is being investigated;
- natural products modulating
known resistance mechanisms (such as, for instance, epicatechin gallate,
a polyphenol extracted from green tea, which sensitizes MRSA to beta-lactams
by delocalizing PBP2 from the septum of dividing bacteria without interacting
with PBP2a [Figure 3B]).
|
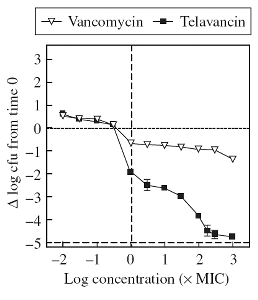
Figure 3A
|
Figure 3
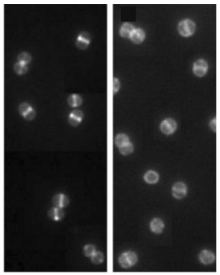
B
|
Figure 3:
Left: Comparative activity of the glycopeptide vancomycin
and of the lipopglycopeptide telavancin. S. aureus was exposed
to increasing concentrations of each of these drugs during 3 h.
The graph shows the change in bacterial counts from the initial
inoculum. telavancin is bactericidal (3 log decrease in colony forming
units [CFU]) as soon as its concentration reaches 10 x its Minimal
Inhibitory Concentration (MIC), while vancomycin never reaches a
bactericidal effect. The mode of action of lipoglycopeptides is
further discussed in the section on drug-membrane
interactions.
From Barcia-Macay et al, 2006
Right: Delocalization
of PBP2 in MRSA as determined by fluorescence microscopy in bacteria
grown in broth containing oxacillin or oxacillin and epicathecin
gallate (ECg). This delocalization (shown by the group of P. Taylor)
is thought to impair the interaction of PBP2 (acting as transglycosylase)
with PBP2a (acting as transpetptidase), which is essential for maintenance
of growth of MRSA in the presence of beta-lactams. This may explain
why ECg sensitizes MRSA to beta-lactams although it does not interact
with PBP2a (shown by our group). These features of ECg-induced phenotype
can be explained by changes in the fluid dynamics of the membrane.
From Bernal et al., 2010
|
|
Selected References
other targets (by reverse
chronological order; for a full reference list, see our publication
list)
- Bernal
P, Lemaire S, Pinho MG, Mobashery S, Hinds J, Taylor PW. 2010. Insertion of
epicatechin gallate into the cytoplasmic membrane of methicillin-resistant
Staphylococcus aureus disrupts penicillin binding protein (PBP) 2a-mediated
beta-lactam resistance by delocalizing PBP2. Journal of Biological Chemistry
(2010) 285:24055-24065 (PDF)
- Franchini C, Muraglia
M, Corbo F, Florio MA, Di Mola A, Rosato A, Matucci R, Nesi M, Van Bambeke
F, Vitali C. 2009. Synthesis and biological evaluation of 2-mercapto-1,3-benzothiazole
derivatives with potential antimicrobial activity Archiv der Pharmazie - Chemistry
in Life Sciences (Weinheim) 342:605-613 (PDF)
- Van
Bambeke F, Mingeot-Leclercq MP, Struelens MJ, Tulkens PM. The bacterial envelope
as a target for novel anti-MRSA antibiotics Trends in Pharmacological Sciences
(2008) 29:124-134 (PDF)
- Barcia-Macay
M, Lemaire S, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. 2006. Evaluation
of the Extracellular and Intracellular Activities (human THP-1 macrophages)
of Telavancin vs. Vancomycin against Methicillin-susceptible, Methicillin-resistant,
Vancomycin-intermediate and Vancomycin-resistant Staphylococcus aureus. Journal
of Antimicrobial Chemotherapy (2006) 58:1177–1184 (PDF)
Expertise
- Study and use of bacterial mutants
with defective growing capabilities
- Target-based and structure-based
drug design for rational discovery and lead optimization of novel antibiotics
- Biochemical and microbiological testing
of new antbiotics for evaluation and characterization of mode of action
Additional information:
<tulkens@facm.ucl.ac.be>
Last significant update: December 2010