|
|
Université catholique
de Louvain (UCL-Bruxelles)
Louvain Drug Research
Institute > Cellular and Molecular Pharmacology |
|
|
Antibiotic
efflux and permeability resistance mechanisms |
|
Quick
links
|
Mutidrug transporters,
present in both bacteria and eucaryotic cells, modulate the accumulation
and disposition of antibiotics, thereby affecting their activity.
In bacteria, we study
the prevalence and expression of efflux pumps in clinical isolates. We
also develop diagnostic tools for the early detection of resistance mediated
by active efflux and by porin alteration.
In eucaryotic cells,
we characterize the efflux transporters of antibiotics in macrophages
and epithelial cells, in relation to the modulation of their intracellular
pharmacokinetics and activity.
These
programs are closely linked to those exploring the chemotherapy
of intracellular infection, drug-membrane
interactions, and the clinical
evaluation of new therapeutic approaches.
|
|
|
|
|
|
|
|
|
 |
| |
Team
- Senior investigators:
F. Van Bambeke, P.M. Tulkens, M.P. Mingeot-Leclercq
- Post-doctoral fellows:
-
- Doctoral fellows:
H. Chalhoub (2013-)
- Former investigators:
J.M. Michot (1998-2003), C. Seral (2002-2003), N. Caceres (2004-2007),
N. Mesaros (2004-2006), L. Avrain (2005-2008), B. Marquez (2005-2010), A.
Lismond (2005-2009 , )S. Carbonnelle (2006-2010), F. El Garch (2006-2009),
M. Riou (2007-2009), Q. Tan (2009-2010),T.T.H. Nguyen (2009-2011), C. Vallet
(2007-2012), J. Buyck (2008-2012), L. Garcia (2008-2012 ), N. Vandevelde
(2010-2014)
Collaborations
- O. Denis, M. Struelens
and F. Jacobs (Hôpital Erasme,
Université libre de Bruxelles, Y. Van Laethem
and A. Dediste (Hôpital Saint-Pierre),
D. Pierard (Universitair Zienkenhuis,
VUB), M. Delmée and A. Simon (cliniques
universitaires St Luc, UCL)
- B. Devreese (Laboratory
for Protein Biochemistry and Biomolecular Engineering; University of
Ghent)
- Y. Glupczynski (Laboratoire
de microbiologie des cliniques
universitaires de l'UCL à Mont Godinne)
- H. Poirel (centre
de Génétique médicale, UCL)
- M. Prévost (Structure
et Fonction des Membranes biologiques, ULB)
- P.
Courvalin (Unité
des Agents antibactériens, Institut Pasteur, Paris, France)
- E. Jacquet (Institut
de Chimie des Substances Naturelles, CNRS, Gif-sur-Yvette, France)
- J.M. Pagès (Perméabilité
et transporteurs membranaires, Université de la Méditerranée,
Marseille, France)
- L. Piddock (Immunity
and Infection, University of Birmingham, Birmingham, UK)
Main current research
programs
General
overview
Amphiphilic
molecules easily cross biomembranes. This has created for cells, probably
very early on in evolution, a need to protect them-selves from inorderly invasion
by these diffusible, potentially harmful molecules, and has resulted in the
emergence of of a large array of efflux pumps with broad substrate specificities.
Because
antibiotics are often amphiphilic, many of them are recognized by these efflux
pumps. Figure 1 shows the topology, the mechanisms of action, and the main
classes of efflux pumps acting on antibiotics which have been described so
far.
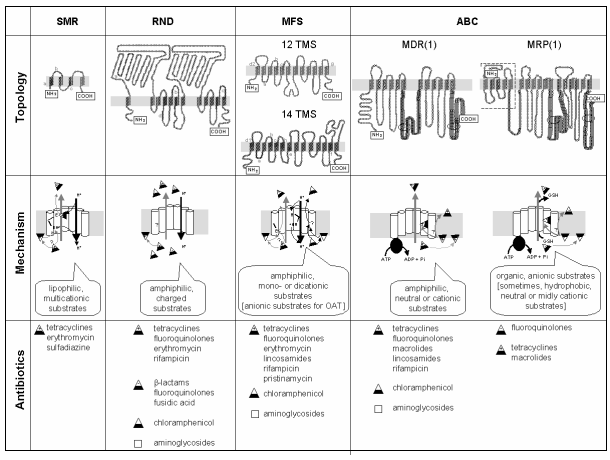 |
Figure 1: Main
classes of efflux pumps acting on antibiotics.
A. within the
class of secondary active transporters (symports, antiports, uniports):
4 superfamilies (comprising at least 10 families of gene products)
including the SMR (Small Multidrug Resistance), the RND
(Resistance Nodulation Division), and the MFS (Major Facilitator
Superfamily).
B. within the class of primary active transporters (energized by ATP):
1 superfamily (ABC) comrpising at last 6 families of gene products
including the PgP (in the MDR1 [Multiple Drug Resistance] group)
and the MRP (Multiple Resistance Protein).
from:
Van Bambeke et al., 2000
|
|
Efflux
in bacteria
Antibiotic efflux pumps are
now recognized as significantly contributing to both innate and acquired bacterial
resistance to many antibiotic because of the very broad variety of substrates
they recognize. Their expression and their cooperation with other mechanisms
of resistance, accounts for so far ill-explained the apparent
intrinsic resistance and "poor susceptibilities"
of important pathogens (e.g. Pseudomonas aeruginosa). Their overexpression
may also explain therapeutic failures, and stable mutations in regulatory
genes could also produce phenotypes of irreversible multidrug resistance.
We evaluate the epidemiology
of efflux-mediated resistance
to current antibiotics in 3 main pathogens (S. aureus, S. pneumoniae, P. aeruginosa)
starting from collections of isolates obtained from clinically-confirmed cases
of respiratory tract and skin and skin
structures infections in Belgium. Efflux
is detected by a combination of
genomic and phenotypic methods. Figure 2 shows a typical example of the detection
of the Mex pumps in Pseudomonas aeruginosa, and the correlation between
the levels of transcription of the genes and the increase in MIC as assessed
by the use of reporter antibiotics.
We also compare the transport
of different antibiotics within a given pharmacological class, with the aim
to delineate to molecular determinants responsible for their recognition by
efflux pumps.
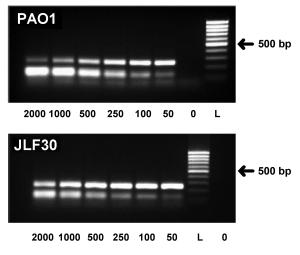
Figure
2:
Genomic detection of Mex pumps in Pseudomonas aeruginosa
and correlation of their level of trannscription and the corresponding
increase in MIC.
(from Mesaros et al.,2007)
|
 |
 |
 |
A:
mexA quantification by QC-RT-PCR in the wild-type PAO1
and in the MexAB-OprM overproducer strain SLF30. Ethidium bromide
stained gels; from left to right: amplification product of cDNA of
the target gene (252 bp) and decreasing amounts (in ag) of the internal
competitor (127 bp).
0 : negative control; L : DNA ladder. |
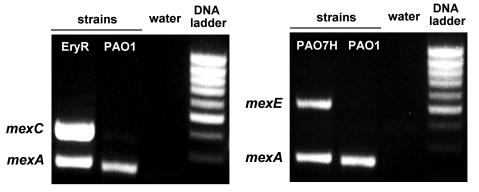
B:
Detection of mexC (left) and mexE (right) expression
by semi-quantitative RT-PCR. The figure shows the ethidium bromide
stained gels of amplified RT-PCR for PAO1 and EryR (left) and for
PAO1 and PAO7H (right). The inducible target genes (mexC, 374bp; mexE,
516bp) are amplified together with the constitutive mexA gene (252bp)
used as positive control. |
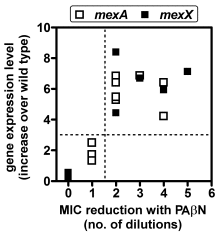
C:
correlation between the level of expression of constitutive MexA and
MexB and the effect of a broad spectrum efflux inhbitor on the MIC
of reporter antibiotics (carbenicillin for mexA and gentamicin
for mexX). |
|
Selected references
on efflux in bacteria (by
reverse chronological order; for a full reference list, see our publication
list)
- Riou M, Avrain L, Carbonnelle
S, El Garch F, Pirnay JP, De Vos D, Plésiat P, Tulkens PM, Van Bambeke
F. Increase of efflux-mediated resistance in Pseudomonas aeruginosa during
antibiotic treatment in patients suffering from nosocomial pneumonia. International
Journal of Antimicrobial Agents (2016) 47:77–83 (PDF)
- Riou M, Carbonnelle S,
Avrain L, Mesaros N, Pirnay JP, Bilocq F, De Vos D, Simon A, Pierard D, Jacobs
F, Dediste A, Tulkens PM, Van Bambeke F, Glupczynski Y. 2010. In vivo development
of antimicrobial resistance in Pseudomonas aeruginosa strains isolated from
lower respiratory tract of intensive care unit patients with nosocomial pneumonia
and receiving antipseudomonal therapy
International Journal of Antimicrobial Agents 36:513-522 (PDF)
- Van Bambeke F, Pagès
J-M, Lee VJ. 2010. Inhibitors of bacterial efflux pumps as adjuvants
in antibacterial therapy and diagnostic tools for detection of resistance
by efflux
In: Frontiers in Anti-Infective Drug Discovery vol. 1, 138-175, Atta-ur-Rahman
& M. Iqbal Choudhary (Eds.), Bentham Science Publishers. Chapter of book.
(PDF)
- El Garch F, Lismond
A, Piddock LJV, Courvalin P, Tulkens PM, Van Bambeke F. 2010. Fluoroquinolones
induce the expression of patA and patB which encode ABC efflux pumps in Streptococcus
pneumoniae. Journal of Antimicrobial Chemotherapy. 65:2076-82
(PDF)
- Mesaros
N, Glupczynski Y, Avrain L, Caceres NE, Tulkens PM, Van Bambeke F. 2007. A
combined phenotypic and genotypic method for the detection of Mex efflux pumps
in Pseudomonas aeruginosa. Journal of Antimicrobial Chemotherapy. 59:378-386.
(PDF)
- Avrain L, Garvey M, Mesaros
N, Glupczynski Y, Mingeot-Leclercq MP, Piddock LJV, Tulkens PM, Vanhoof R,
Van Bambeke F. 2007. Selection of quinolone resistance in Streptococcus pneumoniae
exposed in vitro to sub-inhibitory drug concentrations. Journal of Antimicrobial
Chemotherapy. 60:965-972. (PDF)
- Van Bambeke F.,
Y. Glupczinsky, P. Plésiat, J.C. Pechère & P.M. Tulkens.
2003. Antibiotic efflux pumps in procaryotic cells: impact for resistance
to antibiotic treatments.
J. Antimicrob. Chemother. 51:1055-1065. (Review). (PDF)
Efflux
in eucaryotic cells
Efflux pumps can modulate
the accumulation of antibiotics in phagocytic cells and also play significant
roles in the transepithelial transport of these drugs.
We study the efflux of
antibiotics in phagocytic cells and try to identify, at the phenotypic
and genotypic levels, the transporter(s) involved (Figures 3 A and 3B).
An original approach in
these studies has been the obtention and characterization of macrophages overexpressing
efflux transporters for fluoroquinolone antibiotics (by exposure to progressively
increasing concentrations, so as to obtain 'resistant cells'). We currently
analyze the protein expression of these cells by molecular biology approaches
and proteomics (Figure 3C).
|
|
Figure 3A:
Demonstrating ciprofloxacin efflux in J774 macrophages
Kinetics of accumulation (left) and efflux (right) of ciprofloxacin
(extracellular concentration, 17 mg/liter [50 µM]) in J774
macrophages incubated in the presence or absence of 10 mM probenecid
(the drugs were added simultaneously for the accumulation experiment).
This figure illustrates that the quinolone antibiotic ciprofloxacin
is substrate for a probenecid-inhibitable organic anion transporter,
which we tentitatively identify as a member of the MRP family (see
figure 1). This is compatible with the physicochemical properties
of the drug, which contains a ionizable carboxylic acid.
From Michot
et al. 2004
|
|
|
|
Figure 3B
: Demonstrating the role of P-glycoprotein in reducing the accumulation
of azithromycin in J774 macrophages
Kinetics of accumulation (left) of azithromycin (extracellular concentration,
5 mg/liter) in J774 murine macrophages incubated for up to 24 h
in the absence (open squares) or in the presence (closed squares)
of 20 µM verapamil; kinetics of efflux of azithromycin (extracellular
concentration, 20 mg/liter) from J774 murine macrophages incubated
for 3 h with azithromycin (right).
In this case, the figure illustrates that azithromycin, an amphiphilic
dicationic macrolide antibiotic, is transported by P-glycoprotein,
since its accumulation and efflux can be modulated by the addition
of an inhibitor of this pump.
from Seral et al, 2003
|
|
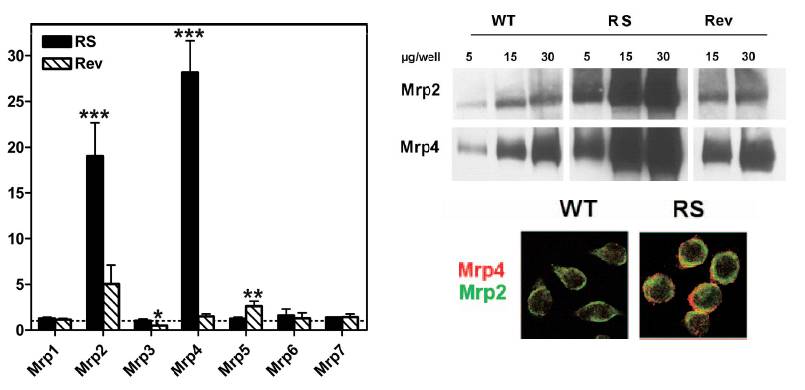
|
Figure 3C: Study of the expression of Mrp
transporters in macrophages made resistant to ciprofloxacin by prolonged
exposure to high concentrations of this drug (RS), as compared to
wild type cells (WT) or revertant cells (obtained by cultivation
of resistance cells in the absence of ciprofloxacin)
Left: Quantification
of mRNA transcripts of Mrps 1 to 7 in ciprofloxacin-resistant (RS)
and revertant (Rev) J774 macrophages in comparison with wild-type
cells (WT). Results are expressed as the increase in expression
over that in WT cells (set arbitrarily at 1 [dotted line]).
Right: Western
Blot revealing Mrp2 or Mrp4 in membrane-ennriched preparation and
colocalization of these transporers at the membrane of WT or RS
cells.
from Marquez
et al, 2009
|
|
In a broader pharmacological
context, we also try
- to delineate structure-activity
relationships for recognition by efflux transporters within a given class
of antibiotics
- to evaluate the consequences
of this efflux for the activity of antibiotics against intracellular bacteria
(see also chemotherapy
of the intracellular infection)
- to identify and characterize
the active transporters involved in the transepithelial movements of antibiotics,
with the aim to explain their absorption and/or distribution in the organism
(see Figure 4).
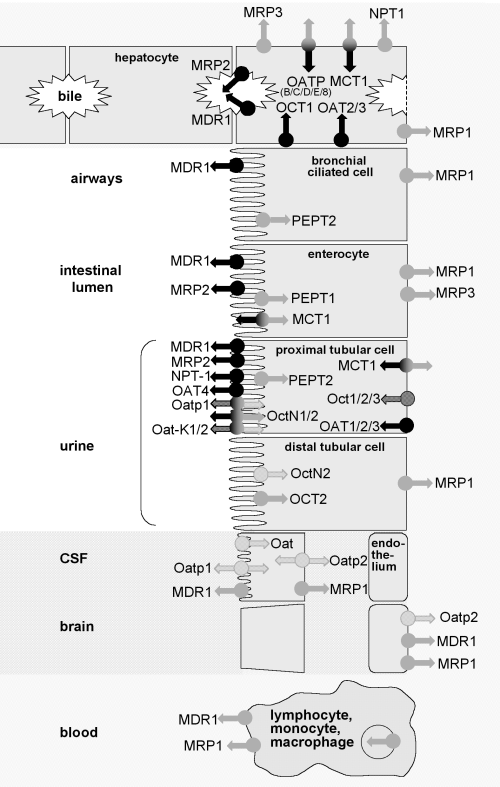
|
Figure
4:
Active transport of antibiotics in eucaryotes
Schematic representation
of the main transporters potentially involved in antibiotic movement
at the level of epithelial cells in the main organs (liver, bronchial
tree, intestine, kidney), the blood-brain barriers, and in leucocytes
(PMN are not considered here since the role of drug transporters
in these cells is unclear).
Black arrows denote transport towards extracorporeal compartments
such as urine, bile, intestine and airways (i.e. transporters involved
in drug elimination from the body). Grey arrows describe uptake
processes from extracorporeal fluids into cells (i.e. allowing drugs
to accumulate in tissues), or from cells to body fluids (i.e. causing
the drug to be transported from one body fluid to another [from
blood to cerebrospinal fluid, e.g.]).
The level of expression of each transporter may differ between species
(arrows with a checkerboard background indicate transporters that
have been, so far, evidenced in animals only). The direction
of transport of bidirectional transporters may differ according
to the cell type.
from: Van
Bambeke et al., 2003. |
|
Selected references
on efflux in eucaryotic cells (by
reverse chronological order; for a full reference list, see our publication
list)
- Marquez B, Pourcelle
V, Vallet CM, Mingeot-Leclercq MP, Tulkens PM, Marchand-Bruynaert J, Van Bambeke
F. Pharmacological characterization of 7-(4-(piperazin-1-yl)) ciprofloxacin
derivatives: antibacterial activity, cellular accumulation, susceptibility
to efflux transporters, and intracellular activity. Pharmaceutical Research
(2014) 31:1290–1301. (PDF)
- Marquez
B, Caceres NE, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. 2009. Identification
of the efflux transporter of the fluoroquinolone antibiotic ciprofloxacin
in murine macrophages: studies with ciprofloxacin-resistant cells.
Antimcrobial Agents and Chemotherapy
(2009) 53: 2410-2416. (PDF)
- Becker JP, Depret G, Van Bambeke
F, Tulkens PM, Prevost M. 2009. Molecular models of human P-glycoprotein in
two different catalytic states. BMC Structural Biology 9:3. (PDF)
- Lismond A, Tulkens PM,
Mingeot-Leclercq MP, Courvalin P, Van Bambeke, F. 2008. Cooperation between
prokaryotic (Lde) and eukaryotic (MRP) efflux transporters in J774 macrophages
infected with Listeria monocytogenes. Studies with ciprofloxacin and moxifloxacin.
Antimicrobial Agents and Chemotherapy 52:3040-3046. (PDF)
- Lemaire S, Van Bambeke
F, Mingeot-Leclercq M-P, Tulkens PM. 2007. Modulation of the Cellular Accumulation
and Intracellular Activity of Daptomycin towards phagocytized Staphylococcus
aureus by the P-glycoprotein (MDR1) Efflux Transporter in human THP-1 macrophages
and Madin-Darby canine kidney cells.Antimicrobial Agents and Chemotherapy
(2007) 51:2748-2757. (PDF)
- Vandevuer S, Van Bambeke
F, Tulkens PM, Prévost M. 2006. Predicting the three-dimensional structure
of human P-glycoprotein in absence of ATP by computational techniques embodying
cross-linking data: insight into the mechanism of ligand migration and binding
sites. Proteins (Proteins-structure function and bioinformatics) (2006) 63:466-478
(Original paper)
(PDF)
- Michot JM, Heremans MF,
Caceres NE, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. 2006. Cellular
accumulation and activity of quinolones in ciprofloxacin-resistant J774 macrophages.
Antimicrobial Agents and Chemotherapy 50:1689-1695 (PDF)
- Michot JM, Seral C, Van
Bambeke F, Mingeot-Leclercq MP, Tulkens PM. 2005. Influence of efflux transporters
on the accumulation and efflux of four quinolones (ciprofloxacin, levofloxacin,
garenoxacin, and moxifloxacin) in J774 macrophages. Antimicrobial Agents and
Chemotherapy 49:2429-2437. (PDF)
- Michot
J-M, F. Van Bambeke, M.P. Mingeot-Leclercq & P.M. Tulkens. 2004.
Active efflux of ciprofloxacin from J774 macrophages through an MRP-like transporter.
Antimicrob. Agents and Chemother. 48:2673-2682. (PDF)
- Seral, C., S. Carryn, P.M. Tulkens,
F. Van Bambeke. 2003. Influence of P-glycoprotein and MRP efflux pump
inhibitors on the intracellular activity of azithromycin and ciprofloxacin
in macrophages infected by Listeria monocytogenes or Staphylococcus aureus.
J. Antimicrob. Chemother. 51:1167-1173. (PDF)
- Seral,
C., J.M. Michot, H. Chanteux, M.P. Mingeot-Leclercq, P.M. Tulkens,F. Van Bambeke.
2003. Influence of P-glycoprotein Inhibitors on the accumulation of
macrolides in J774 murine macrophages. Antimicrob Agents Chemother.
47:1047-1051. (PDF)
- Van Bambeke F., J.M. Michot,
P.M. Tulkens. 2003. Antibiotic efflux pumps in eucaryotic cells: Impact on
antibiotic cellular pharmacokinetics, pharmacodynamics and toxicodynamics.
J. Antimicrob. Chemother. 51:1067-1077. (Review). (PDF)
- F.
Van, Bambeke, E. Balzi, P.M. Tulkens. 2000. Antibiotic efflux pumps (Commentary).
Biochemical Pharmacology 60:457-470. (PDF)
Expertise
and original material
- Study of antibiotic transport
in bacteria (P. aeruginosa, S. pneumoniae, S. aureus) and
development of corresponding diagnostic tools
- original material:
collection of isolates from validated clinical origin and with known expression
of antibiotic efflux transporters
- Study of antibiotic transport
in eucaryotic cells (polarized and non polarized cultures).
- original material:
stable eucaryotic cell lines overexpressing fluoroquinolone efflux pumps
Additional information: <tulkens@facm.ucl.ac.be>
Last significant update: December
2010